Understanding the practical value: Genotyping PRRSv
By Professor Daniel C. L. Linhares
Iowa State University - College of Veterinary Medicine
 The lineage system: How is it created?
The lineage system: How is it created?
In 2023 the lineage system was refined using available global ORF5 sequences. In summary, it classifies PRRSv-2 into 11 distinct lineages. Some lineages are more diverse than others. Thus, they sub-divided Lineage 1 into L1A-J (the most diverse lineage). Within L1C, there are 5 distinct sub-lineages (L1C.1 – L1C.5). Similarly, Lineages 5, 8 and 9 were also classified into sub-lineages.
What are the advantages of the lineage system?
The lineage system based on ORF5 sequencing is able to better characterize the great genetic diversity of PRRSv. This aspect is not fully captured by the older classification method using Restriction Fragment Length Polymorphism (RFLP) pattern-based typing.
What are the disadvantages?
It is still only based on ORF5, which is 4% of the whole genome. Furthermore, it is not useful to predict virulence. There is also no correlation between vaccine efficacy and wild type virus based on similarities in ORF5.
The lineage system: What is the practical value?
- Useful for epidemiological investigations and a helpful tool to understand the genetic relationship between a particular strain and that of neighbors, previous outbreaks and other herds with epidemiologic connections.
- It is beneficial in understanding biosecurity, such as how many unrelated strains are introduced into my herd within a given time frame.
- It can differentiate between wild-type and vaccine-like variants.
How you cannot use it:
- To assess recombination as you need whole genome sequencing (WGS) for that, not only ORF5 sequencing
- To assess point mutations over time or relative to reference strains (need WGS).
- To predict virulence of the strain.
- To assess or predict vaccine efficacy. For that a pig challenge model is needed (gold standard).
How to tell if a PRRS vaccine offers cross protection
a. Pig challenge model for that particular strain. 
b. Not based on phylogenetic tree.
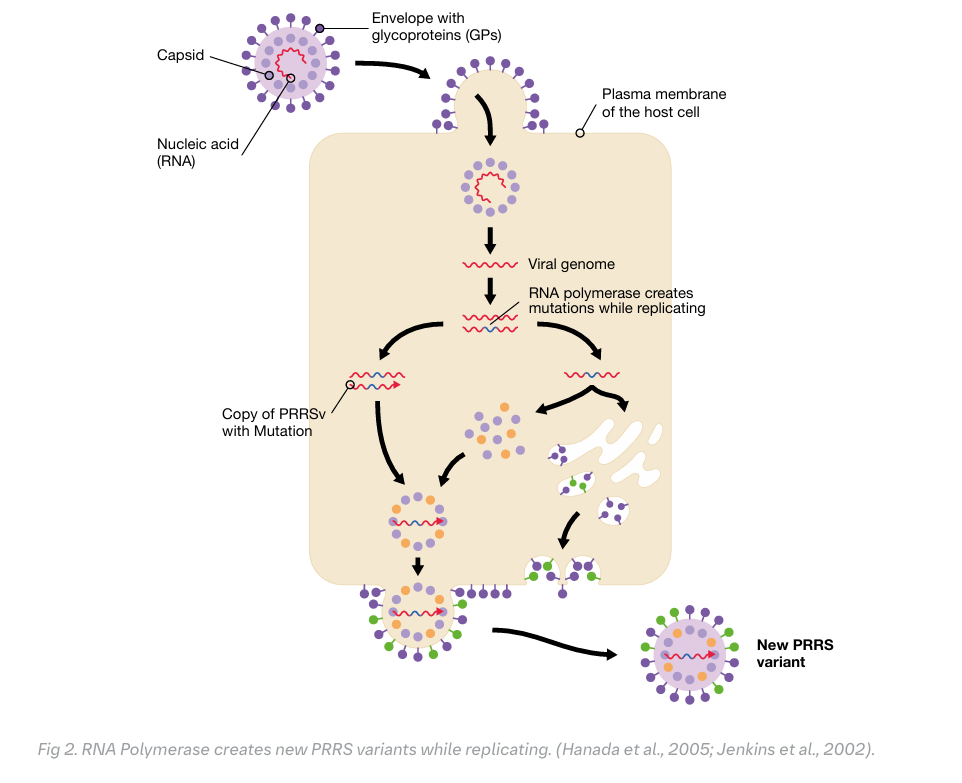
Why is PRRS so diverse?
The enzyme PRRS virus uses for replication is very efficient in producing copies, but lacks a proofreading. So the newly produced viruses are a cloud of variants or mutants resulting in a great diversity. The PRRSv evolution is further accelerated when two different viruses infect a single cell. Therefore, PRRSv is the most diverse virus, resulting in even a greater diversity than human immunodeficiency virus (HIV) and Influenza A virus (IAV). To reflect this huge diversity, PRRSv is clustered in two species (PRRSv-1 and PRRSv-2), lineages and sub-lineages.
You can download the doocument below
Contacto:
Contacta con nosotros a través del siguiente formulario.
Un resumen semanal de las novedades de 3tres3 México
Accede y apúntate a la lista

